Abstract
Established infections with the human and simian immunodeficiency viruses (HIV and SIV, respectively) are thought to be permanent with even the most effective immune responses and antiretroviral therapies only able to control, but not clear, these infections1,2,3,4. Whether the residual virus that maintains these infections is vulnerable to clearance is a question of central importance to the future management of millions of HIV-infected individuals. We recently reported that approximately 50% of rhesus macaques (RM; Macaca mulatta) vaccinated with SIV protein-expressing rhesus cytomegalovirus (RhCMV/SIV) vectors manifest durable, aviraemic control of infection with the highly pathogenic strain SIVmac239 (ref. 5). Here we show that regardless of the route of challenge, RhCMV/SIV vector-elicited immune responses control SIVmac239 after demonstrable lymphatic and haematogenous viral dissemination, and that replication-competent SIV persists in several sites for weeks to months. Over time, however, protected RM lost signs of SIV infection, showing a consistent lack of measurable plasma- or tissue-associated virus using ultrasensitive assays, and a loss of T-cell reactivity to SIV determinants not in the vaccine. Extensive ultrasensitive quantitative PCR and quantitative PCR with reverse transcription analyses of tissues from RhCMV/SIV vector-protected RM necropsied 69–172 weeks after challenge did not detect SIV RNA or DNA sequences above background levels, and replication-competent SIV was not detected in these RM by extensive co-culture analysis of tissues or by adoptive transfer of 60 million haematolymphoid cells to naive RM. These data provide compelling evidence for progressive clearance of a pathogenic lentiviral infection, and suggest that some lentiviral reservoirs may be susceptible to the continuous effector memory T-cell-mediated immune surveillance elicited and maintained by cytomegalovirus vectors.
Similar content being viewed by others
Main
Clinical and experimental observations have suggested that HIV and SIV infections might be vulnerable to immune control or pharmacological clearance in the first few hours to days of infection, before both the viral amplification needed for efficient mutational escape and the establishment of the highly resilient viral reservoir that sustains the infection4,6,7,8. Cytomegalovirus (CMV) vectors were designed to exploit this putative window of vulnerability, based on their ability to elicit and indefinitely maintain high frequency, effector-differentiated, and broadly targeted virus-specific T cells in potential sites of early viral replication5,9,10. Indeed, the pattern of protection observed in approximately 50% of RhCMV/SIV vector-vaccinated RM after intrarectal SIVmac239 challenge was consistent with early immunological interception of the nascent SIV infection at the portal of viral entry and immune control before irreversible systemic spread5. Protected RM manifested a very transient viraemia at the onset of infection, followed by control of plasma SIV levels to below the threshold levels of quantification, except for occasional plasma viral ‘blips’ that waned over time, and after one year, demonstrated only trace levels of tissue-associated SIV RNA and DNA at necropsy using ultrasensitive assays. The occurrence of plasma viral blips and the recurrence of ‘breakthrough’ progressive SIV infection in 1 of the 13 RhCMV/SIV vector-protected RM at day 77 after infection indicated that SIV was not immediately cleared from these protected RM, but the failure to find more than trace levels of SIV nucleic acid in systemic lymphoid tissues was consistent with the productive infection being largely contained at the portal of entry with the possibility of eventual clearance. Given the crucial importance of understanding the degree to which a highly pathogenic lentivirus can be contained or even cleared by adaptive immunity, we sought to define more precisely the spread and dynamics of SIV infection in RM that controlled the infection as a consequence of RhCMV/SIV vector vaccination, and in particular, the extent to which residual SIV was eventually cleared from these animals.
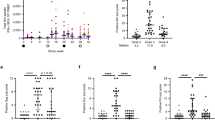
To establish the extent of SIV spread early after the onset of RhCMV/SIV vector-mediated control, we studied a group of five RM vaccinated with RhCMV vectors containing SIV Gag, Rev/Tat/Nef, Env and Pol (but not Vif) inserts that were taken to necropsy within 24 days of controlling plasma viraemia after intrarectal inoculation with SIVmac239. All of these RM had measureable SIV RNA in plasma for one or two weekly time points after challenge, followed by at least three consecutive weekly samples with plasma SIV RNA below 30 copy equivalents (equiv.) per ml, and at the time of necropsy, below 5 copy equiv. ml−1, as measured by an ultrasensitive assay (Fig. 1a). Infection was confirmed by the de novo development of T-cell responses against SIV Vif (not included in the vaccine) in all RM (Fig. 1b and Supplementary Fig. 1a). As previously described5, protection occurred without anamnestic boosting of vaccine-elicited SIV-specific CD8+ T-cell responses in blood (Fig. 1b), and at necropsy, robust CD4+ and CD8+ T-cell responses to the SIV proteins included in the RhCMV/SIV vaccine vectors were identified (Supplementary Fig. 1b). We then used ultrasensitive, nested quantitative PCR (qPCR) and quantitative PCR with reverse transcription (qRT–PCR) assays to quantify SIV DNA and RNA, respectively, in the tissues of these protected RM, in comparison with tissues from three unchallenged, RhCMV/SIV vector-vaccinated RM (SIV− controls), two unvaccinated RM with productive SIV infection (one progressor and one elite controller) and three RM with SIV infection suppressed with antiretroviral drug treatment (ART) (Fig. 1c, Supplementary Figs 2–4 and Supplementary Table 1). Two of the five RhCMV/SIV vector-protected RM showed levels of SIV DNA and RNA approaching the very low level background signal observed for SIV− control RM. However, the other three showed readily measurable SIV RNA, not only in rectal/colonic mucosa (portal of entry), but also in lymph nodes draining the portal of entry (iliosacral and mesenteric lymph node groups), as well as in sites of presumed haematogenous spread: bone marrow, spleen and liver. The level of SIV RNA in the tissues of these three RM was less than that seen in progressive infection, but comparable to that in the elite SIV controller and in ART-suppressed SIV infection. Notably, however, levels of tissue-associated SIV DNA in the RhCMV/SIV vector-protected RM were all substantially lower than in the RM with elite control and ART suppression, probably reflecting virological control before, rather than after, peak viral replication in the RhCMV/SIV vector-protected RM, and the limited time for SIV DNA+ cells to accumulate in these RM before necropsy. Although these data suggest a much smaller SIV reservoir in the RhCMV/SIV vector-protected RM than in the SIV+ controls, including the RM with ART-suppressed SIV infection, we were able to recover replication-competent SIV from iliosacral lymph nodes and spleen in all five of the RhCMV/SIV-protected RM taken to early necropsy (and from bone marrow and mesenteric lymph nodes in three of these five RM), including the two RM with near background levels of SIV RNA by nested qRT–PCR (Table 1). This replication-competent SIV was found in tissues manifesting only minimal interferon-stimulated gene expression, significantly less than found in either progressive or ART-suppressed SIV infection (Supplementary Fig. 5). Taken together, these data demonstrate that in RhCMV/SIV vector-protected RM, SIV can escape the portal of entry and establish infection in draining lymph nodes, as well as bone marrow, spleen and liver, before stringent control.
a, Plasma viral load (measured as log(copy equiv. per ml)) profiles of five RhCMV/SIV vector-vaccinated RM with complete control of viraemia after intrarectal SIVmac239 challenge. All five RM controlled viraemia to below the 30 copy equiv. ml−1 limit of quantification for the standard plasma viral load assay used for all pre-necropsy samples, and to below the 1–5 copy equiv. ml−1 limit of detection for the ultrasensitive plasma viral load assay used on necropsy samples (individual detection limits for each terminal sample shown). b, Frequencies of peripheral blood memory CD8+ T cells specific for SIV proteins that were (Gag plus Pol) or were not (Vif) included in the RhCMV/SIV vectors, shown before and after the onset of the controlled SIV infection. The response frequencies (mean ± s.e.m.) were normalized to the response frequencies immediately before SIV infection for the vaccine-elicited SIV Gag- and Pol-specific responses, and to the peak frequencies after SIV infection for the de novo SIV Vif-specific responses. c, Analysis of tissue-associated SIV DNA and RNA (copy equiv. per 108 cell equiv.) in the five RhCMV/SIV vector-protected RM at necropsy using ultrasensitive quantitative PCR and RT–PCR. BM, bone marrow; LN, lymph nodes; PID, post-infection day; SIM, small intestine mucosa.
After intrarectal inoculation, SIV infection has been reported to spread to draining lymph nodes within 4 h (ref. 11), a rate of dissemination that may preclude SIV-specific effector memory T cells from containing the infection within the mucosa. By contrast, the development of SIV infection after intravaginal inoculation has been reported to require local amplification, with distal spread only after 4–5 days6. To determine whether RhCMV/SIV vector-elicited T-cell responses might locally control and perhaps clear an intravaginal SIV challenge, we compared the outcome of repeated, limiting dose intravaginal SIVmac239 challenge in cycling female RM vaccinated twice (weeks 0 and 14) with RhCMV/SIV vectors (group A) versus similar RM vaccinated twice with RhCMV vectors encoding non-SIV inserts (group B) or left unvaccinated (group C), with challenge 78 weeks after initial vaccination (Supplementary Fig. 6). The immunogenicity of RhCMV/SIV vectors in these female RM was similar to that described for male RM with robust, effector memory-biased SIV-specific CD4+ and CD8+ T-cell responses to all SIV inserts (Supplementary Figs 7 and 8), but little to no SIV Env-specific antibody responses (Supplementary Fig. 9). As previously described for intrarectal SIV challenge of male RM5, RhCMV/SIV vector vaccination did not significantly affect the number of intravaginal SIV challenges required to achieve infection relative to control-vaccinated and unvaccinated RM (Supplementary Fig. 10), but did markedly alter the course of SIV infection with 9 out of 16 RhCMV/SIV vector-vaccinated female RM manifesting stringent (MHC class I allele-independent) control of plasma viraemia compared with none of 18 infected female control RM (Fig. 2a and Supplementary Table 2). Five of these nine protected female RM manifested a second episode of transient plasma viraemia within the first 12 weeks after initial control, but overall, the fraction of protected female RM (followed for at least 30 weeks) with such plasma viral blips (56% versus 100%; P = 0.02 by Fisher’s exact test) and the number of blips per RM (0.7 versus 6.0; P < 0.0001 by two-sided Wilcoxon rank sum test) were less than that observed in RhCMV/SIV vector-vaccinated male RM protected after intrarectal challenge5. Other characteristics of protection in these intravaginally challenged, RhCMV/SIV vector-vaccinated female RM were identical to those previously reported for RhCMV/SIV vector-mediated protection of male RM against intrarectal challenge5, including development of de novo SIV Vif-specific CD4+ and CD8+ T-cell responses, lack of an anamnestic boost of the vaccine-elicited SIV-specific CD4+ or CD8+ T cells, lack of SIV Env seroconversion, and lack of CD4+ T-cell depletion at mucosal effector sites (Fig. 2b and Supplementary Figs 9, 11 and 12).
a, Plasma viral load (measured as log(copy equiv. per ml)) profiles of groups A (RhCMV/SIV vector-vaccinated), B (control RhCMV vector-vaccinated) and C (unvaccinated) RM after infection by repeated, limiting dose, intravaginal SIVmac239 challenge, with the day of infection defined as the challenge before the first above-threshold plasma viral load. The fraction of infected RM that met controller criteria (see Methods) in group A (9 out of 16) versus groups B and C (0 out of 18) was significantly different (P = 0.0002) by two-sided Fisher’s exact test. Note that Rh20363 initially manifested aviraemic protection, but then relapsed with productive, albeit controlled, infection at week 31 after infection. b, Mean (and s.e.m.) frequencies of peripheral blood memory CD8+ T cells specific for SIV proteins that were (Gag plus Pol) or were not (Vif) included in the RhCMV/SIV vectors, measured before and after the onset of SIV infection in the nine group A RM with initial aviraemic control (response frequencies normalized as described in Fig. 1b). Asterisks indicate n = 8 (minus Rh20363 post-relapse); daggers indicate n = 7 (minus Rh20363 and Rh20347, the latter used in the CD8+ cell depletion study described in Supplementary Fig. 14). c, Quantification of tissue-associated SIV RNA (left) and DNA (right) (copies per 108 cell equiv.) in the designated longitudinal samples of the nine group A controllers versus two representative viraemic progressors. All sample types were analysed at weeks 5, 9 and 17 in all RM. All sample types were analysed a fourth time in all controller RM between post-infection weeks 30 and 40, and PBMCs, bone marrow and lymph node samples were analysed a fifth time in eight out of nine controller RM between post-infection weeks 42 and 55. Each symbol represents a single determination from the designated tissue, except when a multiplication factor is shown (for example, ×7 indicates a total of seven samples from different RM with below threshold measurements for that time point).
To determine whether SIV infection spread from the cervical/vaginal mucosa in the nine RhCMV/SIV vector-protected female RM, we biopsied bone marrow, peripheral lymph nodes (axillary/inguinal) and small intestinal mucosa for nested qRT–PCR and qPCR analysis of SIV RNA and DNA, respectively, at 5, 9, 17 and >30 weeks after infection. Notably, in the first 9 weeks of infection, five of these nine RM manifested levels of SIV RNA in bone marrow comparable to levels seen in uncontrolled SIV infection, but, whereas in uncontrolled infection SIV RNA levels were similarly high in peripheral blood mononuclear cells (PBMCs), lymph nodes and intestinal mucosa, SIV RNA was either not detected or detected only at very low levels in these sites in the RhCMV/SIV vector-protected RM (Fig. 2c). Moreover, in contrast to uncontrolled infection, SIV DNA was inconsistently detected in the samples from the RhCMV/SIV vector-protected RM, and by 40 weeks after infection, all nine of the RhCMV/SIV vector-protected RM had at least one sample set in which both SIV RNA and DNA were below the level of detection in all sites. In eight of these RM (excluding Rh20363, see below), all samples obtained subsequent to 30 weeks after infection showed SIV RNA and DNA levels below the level of detection, with the exception of one PBMC sample with low-level SIV RNA (454 copy equiv. per 108 cells). The differences in the frequency of SIV detection in samples obtained at 5, 9 and 17 weeks versus >30 weeks after infection from these eight RM were highly significant (P = 0.002 for all samples, P = 0.0006 for bone marrow by two-sided Wilcoxon rank sum tests).
The ability to detect tissue-associated SIV early, but not late, after infection in these eight stably protected female RM, particularly in bone marrow, is consistent with initial spread and subsequent control and progressive clearance of SIV. In accordance with this, the frequencies of circulating SIV Vif-specific T cells, which are elicited and maintained by antigen derived from SIV infection (rather than the vaccine), progressively declined in these RM until these responses were no longer detectable (Fig. 2b and Supplementary Fig. 11). However, despite having no detectable SIV RNA or DNA in PBMCs and tissue samples at week 17, and declining SIV Vif-specific T-cell responses, one animal (Rh20363) showed the emergence of low-level productive SIV infection at week 31 after infection (Fig. 2a). The boosting of SIV-specific CD4+ and CD8+ T-cell responses (Fig. 2b and Supplementary Fig. 11), including de novo CD8+ T-cell responses to canonical Mamu-A*01-restricted SIV epitopes (Supplementary Fig. 13), the appearance of cell-associated RNA and DNA in subsequent PBMCs, lymph node and intestinal samples (Fig. 2c), and the induction of increased plasma and PBMC-associated SIV loads with experimental in vivo CD8+ lymphocyte depletion (Supplementary Fig. 14) indicates that this RM spontaneously converted from a unique state of stringent viral containment with little or no continuing viral replication to a different state characterized by continuing, but low-level SIV replication (consistent with conventional ‘elite’ immunological control). In keeping with this, sequence analysis of the breakthrough virus 3 weeks after initial viral rebound showed little evolution from the initial SIVmac239 sequence except, notably, a putative escape mutation in the Tat-SL8 epitope sequence, consistent with early escape from the Tat-SL8-specific T-cell responses that developed after viral rebound at week 31 (Supplementary Figs 13 and 15). Given the enormous breadth of RhCMV/SIV vector-elicited CD8+ T-cell responses10, this limited sequence evolution suggests that the loss of aviraemic control in Rh20363 was more likely due to inadequate immune surveillance of residual infection than mutational escape. Experimental CD8+ lymphocyte depletion was also performed on three RhCMV/SIV vector-protected female RM that retained aviraemic control, and in keeping with previous analysis of CD8+ lymphocyte depletion of RhCMV/SIV vector-vaccinated male RM protected after intrarectal challenge5,9, this treatment did not induce detectable plasma viraemia (Supplementary Fig. 14). However, one of these RM (Rh21176) transiently manifested unequivocal detection of SIV RNA (10 out of 10 replicates positive) and replication-competent SIV (7 out of 20 co-cultures positive) in lymph nodes at day 10 after CD8+ lymphocyte depletion, demonstrating the presence of at least local, very low level residual SIV infection in this RM after 52 weeks of stringent control. In contrast to Rh20363, Rh21176 maintained aviraemic control, indicating that this RM's immune system either controlled or eliminated residual foci of SIV replication.
The finding that RhCMV/SIV vector-protected RM are able to control haematogenous SIV dissemination after both intrarectal and intravaginal challenge suggested that the immune responses elicited by these vectors might provide protection even when mucosal surfaces are bypassed. To assess this possibility, we challenged six RhCMV/SIV-vaccinated RM with low dose, intravenous SIVmac239, and found that two of these six RM manifested the same pattern of control observed after mucosal challenge—a transient, low-level viraemia associated with the development of an SIV Vif-specific T-cell response, and detection of SIV RNA in bone marrow (high level) and/or PBMCs (low level) early, but not late, after infection (Supplementary Fig. 16). Taken together, these data indicate that (1) RhCMV/SIV vector-elicited immune responses can mediate protection regardless of the route of SIV challenge, (2) viral control is both local and systemic, and (3) replication-competent SIV can persist in several sites for weeks to months in protected RM (even when aviraemic), but seems to decline over time.
To determine the ultimate fate of residual SIV in RhCMV/SIV vector-protected RM, we followed a total of ten protected RM for 69–180 weeks after infection (Fig. 3a, b). In all of these RM, plasma viral blips became increasingly infrequent over time, with no blips observed after 70 weeks. The frequency of the SIV infection-dependent, SIV Vif-specific CD8+ T cells in blood also progressively declined in all RM until these responses were no longer detectable (Supplementary Fig. 17). In contrast to the SIV Vif-specific CD8+ T-cell responses, the SIV-specific CD8+ T-cell responses elicited by the RhCMV/SIV vectors remained stable, including high frequencies of CD8+ T cells capable of recognizing autologous SIV-infected CD4+ T cells (Supplementary Fig. 18). Analysis of six of these medium- to long-term protected RM at necropsy, including one RM that was CD8+ lymphocyte-depleted 10 days before necropsy (Supplementary Fig. 19), confirmed the systemic loss of SIV Vif-specific T cells, and the maintenance of RhCMV vector-elicited, SIV-specific T cells (Supplementary Fig. 20). Most importantly, ultrasensitive, nested qRT–PCR and qPCR analysis of ≥54 tissues per animal (ten replicates per tissue, including extensive sampling of all tissues shown to contain SIV in the short-term RhCMV/SIV vector-protected RM) revealed extremely low to absent levels of SIV RNA and DNA that were indistinguishable from measurements in unchallenged RhCMV/SIV-vaccinated (SIV−) controls (Fig. 3c, d, Supplementary Figs 2 and 21 and Supplementary Tables 1 and 3). Moreover, despite extensive sampling (>240 cultures per animal), no replication-competent SIV was isolated by co-culture analysis from the lymphoid tissues of these RM (Table 1). Finally, we asked whether the adoptive transfer of a total of 6 × 107 haematolymphoid cells (3 × 107 each of peripheral blood leukocytes and lymph node cells, or 3 × 107 each of bone marrow leukocytes and spleen cells) from three SIV+ control RM (two with ART-suppressed infection and one elite controller), and five medium- or long-term RhCMV/SIV vector-protected RM (including one RM tested before and after CD8+ cell depletion) would initiate infection in SIV-naive RM. Remarkably, although cells from the SIV+ controls, including ART-suppressed RM, rapidly initiated SIV infection in the SIV-naive recipients (manifested by the onset of SIV replication and induction of SIV Vif-specific T-cell responses), no evidence of SIV infection was observed in the SIV-naive recipients receiving cells from the medium- and long-term RhCMV/SIV vector-protected RM (Fig. 3e and Supplementary Fig. 22). Taken together, these data provide strong evidence that after being unequivocally infected with SIV, these RhCMV/SIV vector-vaccinated RM cleared detectable infection, such that by all measured criteria (lack of plasma viral blips, absence of Vif-specific T-cell responses, extensive ultrasensitive qRT–PCR and qPCR and co-culture analysis, and adoptive transfer) these RM were indistinguishable from RhCMV/SIV vector-vaccinated controls that had never been exposed to SIV. Although we cannot rule out residual virus below our level of detectability, or in tissues not examined, these data strongly support progressive immune-mediated clearance of an established lentivirus infection, leading to a situation meeting criteria for a functional cure12 and consistent with possible viral eradication.
a, b, Plasma viral load (measured as log(copy equiv. per ml)) profiles of ten RhCMV/SIV vector-vaccinated RM that controlled SIV infection after intrarectal challenge (eight long term (a) and two medium term (b)). The limit of detection for all pre-terminal plasma viral load assays is 30 copy equiv. ml−1; the limit of detection for the ultrasensitive assay used on the terminal sample of the study was ≤1 copy equiv. ml−1. Note that one of the RM with medium-term protection (Rh26467) was CD8+ lymphocyte-depleted 10 days before the terminal sample. c, d, Quantification of tissue-associated SIV DNA and RNA in four long-term and two medium-term protected RhCMV/SIV-vaccinated RM studied at necropsy, including the CD8+ cell-depleted RM (Rh26467). e, Assessment of residual replication-competent, cell-associated SIV in medium- and long-term protected RM by adoptive transfer of 6 × 107 haematolymphoid cells (3 × 107 blood leukocytes and 3 × 107 lymph node cells or, in one transfer from Rh26467, represented by the open symbol, 3 × 107 bone marrow leukocytes and 3 × 107 spleen cells) to SIV-naive RM with SIV infection in the recipient RM delineated by plasma viral load. Cell transfers from RM with conventional elite SIV control and ART-suppressed SIV infection resulted in rapid onset of SIV infection in the recipient RM, but no SIV infection was observed in RM receiving cells from medium- to long-term RhCMV/SIV vector-protected RM (including Rh26467, analysed both before and after CD8+ cell depletion).
In the past 5 years, the HIV/AIDS vaccine field has concluded that a prophylactic HIV/AIDS vaccine must prevent or eliminate HIV infection, as it is thought that any residual infection runs a high risk of eventual progression13. Our demonstration here that the virus-specific, effector memory T cells maintained by a persistent vector can shut down productive SIV infection, and by maintaining immune surveillance over time, functionally cure and possibly eradicate this infection, indicates that an effector memory T-cell-targeted HIV/AIDS vaccine could (by itself, or combined with antibody-targeted approaches) provide meaningful long-term efficacy. Our results also suggest that an effector memory T-cell-targeted vaccine might contribute to HIV cure strategies. Although the SIV reservoirs that initially develop in RhCMV/SIV vector-vaccinated controllers are smaller in size, and possibly different in character from HIV/SIV reservoirs in the setting of ART administration initiated in chronic infection, it is conceivable that the indefinitely persistent, unconventionally targeted10, viral-specific T cells elicited and maintained by CMV vectors—alone or in combination with agents designed to activate HIV gene expression1,2,12—might exert potent immune pressure on cells with any HIV protein expression (including expression of viral antigen by stochastically activated, latently infected cells) and thereby facilitate depletion of residual HIV reservoirs in patients on suppressive ART. It is also possible that these responses might stringently control recrudescent ‘rebound’ infection after ART withdrawal in a manner analogous to their control of primary SIV infection in this study. In summary, the ability of CMV vectors to implement continuous, long-term, and potent antipathogen immune surveillance makes them promising candidates for vaccine strategies intended to prevent and cure HIV/AIDS, as well as other chronic infections.
Methods Summary
RhCMV/SIV vectors expressing SIV Gag, Rev/Tat/Nef, Pol and Env or irrelevant control inserts were used as described5,9. RM were challenged with SIVmac239 by intrarectal, intravaginal or intravenous inoculation using a repeated (weekly), limiting dose protocol5,9. RM were considered infected after detection of plasma SIV RNA ≥ 30 copy equiv. ml−1 and the de novo development of T-cell responses to SIV Vif, an SIV antigen not included in the RhCMV/SIV vectors. Selected RM were depleted of CD8+ lymphocytes, as described14. SIV-specific T-cell responses were measured in mononuclear cells from blood and tissues by flow cytometric intracellular cytokine analysis5,9. Levels of SIV RNA and DNA in tissue/cell preparations were quantified, respectively, by an ultrasensitive, nested qRT–PCR and qPCR approach5. Replication-competent SIV was detected in mononuclear cells by inductive co-cultivation assays, as described15. To ascertain the presence of occult replication-competent SIV below the level of detection of the co-cultivation assay, 60 million cells from blood, lymph nodes, bone marrow or spleen from RhCMV vector-protected RM or RM with spontaneously controlled or ART-suppressed SIV infection were adoptively transferred by intravenous infusion to SIV-naive RM, with infection in the recipient RM determined as described above.
Online Methods
Rhesus macaques
Ninety-nine purpose-bred male and female RM (M. mulatta) of Indian genetic background were used with the approval of the Oregon National Primate Research Center Animal Care and Use Committee, under the standards of the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. These animals were specific pathogen-free as defined by being free of cercopithicine herpesvirus 1, D-type simian retrovirus, simian T-lymphotrophic virus type 1, rhesus rhadinovirus, and Mycobacterium tuberculosis. MHC-1 genotyping for common Mamu alleles such as Mamu-A*01/-A*02 and Mamu-B*08/-B*17 was performed by sequence-specific priming PCR, as described16. The 99 RM include 15 RhCMV/SIV vector-vaccinated RM (7–10 years of age) with aviraemic control of SIV infection (14 with SIVmac239, 1 with SIVmac251) after intrarectal challenge (5 with short-term follow up; 10 with medium- to long-term follow up); 52 RM (4–19 years of age) vaccinated with RhCMV/SIV or control RhCMV vectors or left unvaccinated before SIVmac239 challenge (42 intravaginal, 10 intravenous); 12 SIV+ RM (5–12 years of age; 8 with progressive SIVmac239 infection, 1 with spontaneously controlled SIVmac239 infection, 3 with ART-suppressed SIVmac251 infection); and 20 SIV-naive RM (4–14 years of age) used as negative controls (these include 4 RM vaccinated with RhCMV/SIV vectors) or as SIV- and RhCMV vector-naive recipients in the adoptive transfer experiments. Early (<1 year) follow-up of eight of the RhCMV/SIV vector-vaccinated RM with long-term, aviraemic control of SIV infection was previously reported5, with this study extending that follow-up from 1 to >3 years. RhCMV/Gag, Rev/Tat/Nef, Env and Pol-1 and Pol-2 vectors were administered subcutaneously at a dose of 5 × 106 plaque forming units per vector. The control antigen-expressing RhCMV vector was used at a total dose of 2.5 × 106 plaque forming units to match the total dose of the RhCMV/SIV vectors. RM were vaccinated twice with RhCMV vectors, 14 weeks apart. All SIV challenges (intrarectal, intravaginal and intravenous) used a repeated limiting dose protocol with dosing designed to require >1 challenge for infection of >60% of challenged RM, and to infect all or nearly all challenged RM with ≤10 (weekly) challenges for intrarectal or intravaginal inoculation (300 focus-forming units) and ≤3 (every third week) challenges for intravenous inoculation (0.2 focus-forming units). RM were considered SIV-infected (and challenge discontinued) with the onset of plasma viral load ≥ 30 copy equiv. ml−1 and the de novo development of CD4+ and CD8+ T-cell responses to SIV Vif, an SIV antigen not included in the RhCMV/SIV vectors. RM were considered controllers if plasma viral load became undetectable (<30 copy equiv. ml−1) within 2 weeks of the initial positive plasma viral load and was then maintained below threshold for three consecutive determinations. RM with progressive SIV infection were followed for 20 weeks after infection, or if progression was rapid, until the onset of AIDS. ARTs consisted of two reverse transcriptase inhibitors (20 mg day−1 tenofovir, 50 mg day−1 emtricitabine), an integrase inhibitor (240 mg day−1 raltegravir) and a protease inhibitor (600 mg twice daily darunavir boosted with 100 mg twice daily ritonavir). Selected RM were depleted of CD8+ lymphocytes by administration of 10, 5, 5 and 5 mg per kg body weight of the CD8α monoclonal antibody M-T807R1, a modified version of the cM-T807 humanized anti-CD8 monoclonal antibody with rhesus constant and variable framework regions (http://nhpreagents.bidmc.harvard.edu), administered on days 0, 3, 7 and 10, respectively14. Tissues obtained by biopsy or at necropsy were processed for mononuclear cell preparation, virological and/or immunohistological analysis as previously described5,17. For adoptive transfer experiments, freshly obtained peripheral blood and bone marrow buffy coats were prepared by centrifugation (400g for 20 min). These buffy coats and/or freshly obtained whole lymph node cell or splenocyte preparations were washed three times in saline before intravenous infusion, with each RM receiving 3 × 107 peripheral blood leukocytes plus 3 × 107 lymph node cells (or in 1 RM, 3 × 107 bone marrow leukocytes plus 3 × 107 splenocytes) over 1 h.
Vectors and viruses
The construction and characterization of the strain 68-1-derived RhCMV/SIV vectors, including RhCMV(Gag), RhCMV(Rev/Tat/Nef), RhCMV(Env) and RhCMV(Pol-1) and RhCMV(Pol-2), have been previously described5,9. A control RhCMV vector expressing an M. tuberculosis Ag85B-ESAT6 fusion protein under the control of the EF1-α promoter was constructed with the same E/T recombination approach and RhCMV (68-1) bacterial artificial chromosome used for RhCMV/SIV construction9. RhCMV vector stocks were titred using primary rhesus fibroblasts in a tissue culture infectious dose 50 (TCID50) assay. The pathogenic SIV challenge stocks used in these experiments were generated by expanding the SIVmac239 clone (or SIVmac251 swarm) in RM PBMCs, and were titred using the CMMT-CD4-LTR-β-Gal (sMAGI) cell assay (NIH AIDS Reagent Program).
Viral detection assays
Plasma viral loads were determined by quantitative RT–PCR as previously described18,19. Ultrasensitive determinations of plasma viral loads at necropsy were achieved by concentrating virus from the larger volumes of material available by ultracentrifugation (6041 10-ml tubes and 4018 crown assembly, Seton Scientific; T1270 rotor, Thermo-Sorvall Scientific) at 170,000g for 30 min before processing RNA. Reactions were also run in triplicate and followed the analysis recommendations described previously20 permitting per reaction determinations of 1 copy (2 of 3 positive amplifications) and threshold sensitivities correspondingly lower, dependent on the amount of plasma input. Plasma viral sequencing of Rh20363 post-rebound was performed by synthesis of complementary DNA with SIV gene-specific primers followed by sequencing using the single genome amplification strategy21 (GenBank accessions KF439057, KF439058 and KF439059). Quantitative assessment of SIV DNA and RNA in isolated cells and tissues was determined by the quantitative hybrid real-time/digital RT–PCR and PCR assays, essentially as previously described5, but with modifications to allow more efficient processing of samples larger than ∼100 mg and to increase sample throughput. With respect to the former, the large tissue samples were directly disrupted in TriReagent (Molecular Research Center) using two 11-mm stainless steel balls over 10–15 stainless steel hex nuts (5.5-mm wide) as grinding media, rather than first attempting to cryogenically pulverize the tissue. With respect to the latter, the RT–PCR and PCR assay conditions were also modified to reduce reaction volumes to allow use of 384-well plates. The cDNA reactions were reduced to 15 μl, comprising 10 μl sample plus 5 μl concentrated reaction cocktail and contained 2 mM SIVnestR01 primer, 10 U RNAsin, and 50 U MoMLV reverse transcriptase (Promega). The cDNA synthesis stage of the thermal profile was optimal for MoMLV reverse transcription at 37 °C for 60 min, as opposed to 42°C for 40 min. The RT–PCR pre-amplification reactions were 25 μl in volume with 1.25 U PlatinumTaq polymerase (Life Technologies) and 2.5 μl of this reaction was transferred to 20 μl of real-time PCR reaction mix with 1 U PlatinumTaq polymerase. For DNA determinations, the preamplification reactions were 20 μl in volume, comprising 10 μl sample and 10 μl reaction cocktail; 2 μl of this ‘nested’ reaction was transferred to 20 μl of real-time PCR reaction mix. As previously described, for both RNA and DNA determinations, 12 replicate reactions were tested per sample including a spike of RNA or DNA internal control sequence standard in two of the twelve reactions to assess overall amplification efficiency and assess potential inhibition of the PCR or RT–PCR reactions. The amount of DNA or RNA standard added to replicate reactions to monitor inhibition and PCR performance was typically 10–100 copies, depending on the anticipated level of SIV sequences present. Samples showing greater than a five cycle shift in amplification of the spiked standard, compared to amplification in the absence of specimen nucleic acid, corresponding to less than 74% overall amplification efficiency, were diluted and re-assayed. Quantitative determinations for samples showing amplification in all replicates were derived directly with reference to a standard curve. Quantitative determinations for samples showing fewer than 10 positive amplifications in replicates were derived from the frequency of positive amplifications, corresponding to the presence of at least one target copy in a reaction, according to a Poisson distribution of a given median copy number per reaction. To avoid false positives in biopsy material, in which the specimen size and total number of specimens is limited, we required a minimum of two positive reactions out of ten for a sample to be considered positive. The presence of inducible, replication-competent SIV in mononuclear cell preparations derived from different tissue sites at necropsy was detected by co-cultivation of 2.5 × 105 unfractionated cells from each tissue with 2 × 105 CEMx174 cells (×20 replicates per tissue, cell numbers permitting; CEMx174 cells obtained from NIH AIDS Research & Reference Reagent Program)15. After 18 days, each culture was stained for CD3, CD4 and intracellular SIVgag-p27 (monoclonal antibody 55-2F12), with positive cultures based on ≥0.5% CEMx174 cells with intracellular SIV Gag expression over background by flow cytometry.
Immunological assays
SIV-specific CD4+ and CD8+ T-cell responses were measured in blood and tissues by flow cytometric intracellular cytokine analysis, as previously described in detail5,9. To determine T-cell responses to SIV peptide mixes or individual peptides, mononuclear cells were incubated with mixes of overlapping 15-amino-acid peptides comprising SIV proteins or individual epitopic 8- to 10-amino-acid peptides (with every individual peptide always at 2 μg ml−1) and the co-stimulatory molecules CD28 and CD49d (BD Biosciences) for 1 h, followed by addition of brefeldin A (Sigma-Aldrich) for an additional 8 h. Co-stimulation without antigen served as a background control. To determine responses to autologous SIV-infected cells, SIV+ and SIV− target cells were produced by spinoculating (or not spinoculating) activated CD4+ T cells with sucrose-purified SIVmac239, followed by culturing the cells for 4 days and then purifying the CD4+ cells with CD4 microbeads and LS columns (Miltenyi Biotec), as described22. These cell preparations were >95% CD4+ T cells and the SIV-infected preparations were >50% SIV+ following enrichment. SIV+ versus SIV− T cells were then incubated with microbead-purified CD8+ T cells at an effector:target ratio of 40:1 under the same conditions used for peptide-specific flow cytometric intracellular cytokine analysis. After incubation, stimulated cells were stored at 4 °C until staining with combinations of fluorochrome-conjugated monoclonal antibodies including: SP34-2 (CD3; Pacific Blue, PerCP-Cy5.5), L200 (CD4; AmCyan), SK-1 (CD8α; APC, PerCP-Cy5.5), CD28.2 (CD28; PE, PE-TexasRed), DX2 (CD95; APC, PE), 15053 (CCR7; Pacific Blue), B56 (Ki-67; FITC), MAB11 (TNF-α; APC, FITC, PE), B27 (IFN-γ; APC, FITC) and FN50 (CD69; PE, PE-TexasRed). Data was collected on an LSR-II (BD Biosciences). Analysis was performed using FlowJo software (Tree Star). In all analyses, gating on the lymphocyte population was followed by the separation of the CD3+ T-cell subset and progressive gating on CD4+ and CD8+ T-cell subsets. Antigen-responding cells in both CD4+ and CD8+ T-cell populations were determined by their intracellular expression of CD69 and either or both of the IFN-γ and TNF cytokines. After subtracting background, the raw response frequencies were memory corrected, as previously described5,9. In selected experiments, cells responding to SIV peptides by production of either or both of IFN-γ and TNF were directly phenotyped with respect to the memory markers CD28 and CCR7 (refs 5, 9). Titres of SIV Env-specific antibodies were determined by neutralization of tissue culture-adapted SIVmac251 using a luciferase reporter gene assay23.
Immunohistology
Immunohistochemistry was performed using a biotin-free polymer approach (Golden Bridge International) on 5-μm tissue sections mounted on glass slides, which were dewaxed and rehydrated with double-distilled H2O. Heat-induced epitope retrieval was performed by heating sections in 0.01% citraconic anhydride containing 0.05% Tween-20 in a pressure cooker set at 122–125 °C for 30 s. Slides were incubated with blocking buffer (TBS with 0.05% Tween-20 and 0.5% casein) for 10 min. For APOBEC3G, slides were incubated with rabbit anti-APOBEC3G (1:100; Sigma HPA001812) diluted in blocking buffer overnight at 4 °C. Slides were washed in 1× TBS with 0.05% Tween-20, endogenous peroxidases blocked using 1.5% (v/v) H2O2 in TBS, pH 7.4, for 10 min, incubated with rabbit polink-2 horseradish peroxidase (HRP) and developed with impact DAB (3,3′-diaminobenzidine; Vector Laboratories). For ISG15 staining, after the heat-induced epitope retrieval step, the slides were loaded on an IntelliPATH autostainer (Biocare Medical) and stained with optimal conditions determined empirically that consisted of a blocking step using blocking buffer (TBS with 0.05% Tween-20 and 0.5% casein) for 10 min and an endogenous peroxidase block using 1.5% (v/v) H2O2 in TBS, pH 7.4, for 10 min. Rabbit anti-ISG (1:250; Sigma HPA004627) was diluted in blocking buffer and incubated for 1 h at room temperature. Tissue sections were washed and developed using the Rabbit Polink-1 HRP staining system (Golden Bridge International) according to manufacturer’s recommendations. Sections were developed with impact DAB (Vector Laboratories). All slides were washed in H2O, counterstained with haematoxylin, mounted in Permount (Fisher Scientific), and scanned at high magnification (×200) using the ScanScope CS System (Aperio Technologies), yielding high-resolution data from the entire tissue section. Representative regions of interest (500 mm2) were identified and high-resolution images extracted from these whole-tissue scans. The percentage area of the T-cell zone that stained for APOBEC3G and ISG15 was quantified using Photoshop CS5 and Fovea tools.
Statistical analysis
The RM used in the vaccine efficacy analysis were randomly assigned to vaccine groups with randomization stratified to balance groups for expression of protective MHC alleles. All reported experiments were conducted once and are reported fully. The criteria for categorizing post-challenge RM into ‘protected’ and ‘unprotected’ groups were established previously5. Experimenters were not explicitly blinded to the treatment assignments of the RM, nor were the analyses conducted by blinded investigators. All statistical analyses were conducted using non-parametric and model-independent analysis procedures either for the main analysis or as a sensitivity analysis, and in every sensitivity analysis the result was qualitatively consistent with the main analysis. No tests depended on an assumption of equal variance across compared groups. The only exceptions were the time series analyses, which were conducted with two model-based (regression) approaches; the residuals of these analyses were evaluated and found to be consistent with homoschedasticity and normality requirements, and the results were consistent across approaches. For comparisons of continuous-valued data from independent samples, we applied bivariate Mann–Whitney U tests24, also known as Wilcoxon rank sum tests. For comparisons of dichotomous values across groups, we applied Fisher’s exact tests25. We estimated confidence bounds for binomial proportions using the Wilson score method, as described in Agresti and Coull26. We compared group means of positivity frequencies for which we have repeated binary measures on individual RM using mixed effects logistic regression (with individual RM mean deviations from group means modelled as a normally-distributed random effect). We compared confidence intervals for RM groups to confidence intervals for individual RM (from other groups) by directly determining overlap of the intervals (and we used the estimated random effect variance and estimated group means to conduct z-tests in sensitivity analyses, which yielded consistent results). We compared Kaplan–Meier curves using the log-rank test27. We conducted time series analyses using standard linear regression with time as the primary predictor, and we used Gaussian first-order autoregressive process models in sensitivity analyses, which yielded consistent results. All tests were conducted as two-tailed tests with a type-I error rate of 5%. We used the R statistical computing language28 for all statistical analyses.
Change history
29 October 2014
A Correction to this paper has been published: https://doi.org/10.1038/nature13840
References
Chun, T. W. & Fauci, A. S. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS 26, 1261–1268 (2012)
Lewin, S. R., Evans, V. A., Elliott, J. H., Spire, B. & Chomont, N. Finding a cure for HIV: will it ever be achievable? J. Int. AIDS Soc. 14, 4 (2011)
Deeks, S. G. & Walker, B. D. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27, 406–416 (2007)
Picker, L. J., Hansen, S. G. & Lifson, J. D. New paradigms for HIV/AIDS vaccine development. Annu. Rev. Med. 63, 95–111 (2012)
Hansen, S. G. et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011)
Haase, A. T. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu. Rev. Med. 62, 127–139 (2011)
Lifson, J. D. et al. Containment of simian immunodeficiency virus infection: cellular immune responses and protection from rechallenge following transient postinoculation antiretroviral treatment. J. Virol. 74, 2584–2593 (2000)
Sáez-Cirión, A. et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of Early Initiated Antiretroviral Therapy ANRS VISCONTI Study. PLoS Pathog. 9, e1003211 (2013)
Hansen, S. G. et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nature Med. 15, 293–299 (2009)
Hansen, S. G. et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 (2013)
Ribeiro dos Santos, P. et al. Rapid dissemination of SIV follows multisite entry after rectal inoculation. PLoS ONE 6, e19493 (2011)
Deeks, S. G. et al. Towards an HIV cure: a global scientific strategy. Nature Rev. Immunol. 12, 607–614 (2012)
Burton, D. R. et al. A blueprint for HIV vaccine discovery. Cell Host Microbe 12, 396–407 (2012)
Okoye, A. et al. Profound CD4+/CCR5+ T cell expansion is induced by CD8+ lymphocyte depletion but does not account for accelerated SIV pathogenesis. J. Exp. Med. 206, 1575–1588 (2009)
Shen, A. et al. Novel pathway for induction of latent virus from resting CD4+ T cells in the simian immunodeficiency virus/macaque model of human immunodeficiency virus type 1 latency. J. Virol. 81, 1660–1670 (2007)
Loffredo, J. T. et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81, 8827–8832 (2007)
Tabb, B. et al. Reduced inflammation and lymphoid tissue immunopathology in rhesus macaques receiving anti-tumor necrosis factor treatment during primary simian immunodeficiency virus infection. J. Infect. Dis. 207, 880–892 (2013)
Cline, A. N., Bess, J. W., Piatak, M., Jr & Lifson, J. D. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34, 303–312 (2005)
Venneti, S. et al. Longitudinal in vivo positron emission tomography imaging of infected and activated brain macrophages in a macaque model of human immunodeficiency virus encephalitis correlates with central and peripheral markers of encephalitis and areas of synaptic degeneration. Am. J. Pathol. 172, 1603–1616 (2008)
Palmer, S. et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41, 4531–4536 (2003)
Keele, B. F. et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206, 1117–1134 (2009)
Sacha, J. B. & Watkins, D. I. Synchronous infection of SIV and HIV in vitro for virology, immunology and vaccine-related studies. Nature Protocols 5, 239–246 (2010)
Montefiori, D. C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12, Unit 12.11. (2005)
Wolfe, D. A. & Hollander, M. Nonparametric Statistical Methods (Wiley, 1973)
Fisher, R. A. The logic of inductive inference. J. Roy. Stat. Soc. A 98, 39–54 (1935)
Agresti, A. & Coull, B. A. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52, 119–126 (1998)
Harrington, D. P. & Fleming, T. R. A class of rank test procedures for censored survival data. Biometrika 69, 553–566 (1982)
R Development Core Team. R, A Language and Environment for Statistical Computing; http://www.R-project.org (2011)
Acknowledgements
This work was supported by the AIDS Vaccine Research in Nonhuman Primates Consortium of the National Institute of Allergy and Infectious Diseases (NIAID; U19 AI095985), as well as other NIAID grants (RO1 AI060392, R37 AI054292, P01 AI094417 and U19 AI096109); the Bill & Melinda Gates Foundation-supported Collaboration for AIDS Vaccine Discovery (CAVD); the International AIDS Vaccine Initiative (IAVI) and its donors, particularly the US Agency for International Development (USAID); the National Center for Research Resources (P51 OD011092); and the National Cancer Institute (contract HHSN261200800001E). We thank A. Sylwester, A. Okoye, C. Kahl, S. Hagen, R. Lum, Y. Fukazawa, S. Shiigi and L. Boshears for technical or administrative assistance; C. Miller, N. Miller and T. Friedrich for provision of SIV stocks; K. Reimann for provision of the CD8-depleting monoclonal antibody; D. Watkins for MHC typing; D. Montefiori for neutralizing antibody assays; and A. Townsend for figure preparation. We acknowledge the contribution of M. A. Jarvis to the design, construction and initial in vitro characterization of all the strain 68.1-derived RhCMV vectors used in this study, including both previously published RhCMV/SIV vectors and a previously unpublished vector expressing an Mycobacterium tuberculosis Ag85B–ESAT6 fusion protein, used as a negative control in this study.
Author information
Authors and Affiliations
Contributions
S.G.H. planned and performed animal experiments, and analysed immunological and virological data, assisted by A.B.V., C.M.H., R.M.G., J.C.F., M.S.L., A.N.G., G.X. and N.W. M.P. and J.D.L. planned and performed SIV quantification, assisted by K.O., R.S. and Y.L. B.J.B. and J.B.S. performed infected cell recognition assays. B.F.K. performed sequencing analysis. J.D.E. performed immunohistological studies. S.L.P., J.M.T., A.W.L. and M.K.A. managed the animal protocols. J.A.N. and K.F. supervised CMV vector design and development. J.D.L. planned and supervised SIV quantification and immunohistological experiments. P.T.E. performed all statistical analyses. L.J.P. conceived the RhCMV vector strategy, supervised all experiments, analysed data, and wrote the paper, assisted by S.G.H., J.D.L. and J.B.S.
Corresponding authors
Ethics declarations
Competing interests
Oregon Health & Science University, and authors L.J.P., S.G.H., K.F. and J.A.N. have a considerable financial interest in TomegaVax, Inc., a company that may have a commercial interest in the results of this research and technology. The potential individual and institutional conflicts of interest have been reviewed and managed by Oregon Health & Science University.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-22, Supplementary Tables 1-3 and additional references. (PDF 2610 kb)
Rights and permissions
About this article
Cite this article
Hansen, S., Jr, M., Ventura, A. et al. Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013). https://doi.org/10.1038/nature12519
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12519
This article is cited by
-
Maternal antibodies induced by a live attenuated vaccine protect neonatal mice from cytomegalovirus
npj Vaccines (2023)
-
Cryptic MHC-E epitope from influenza elicits a potent cytolytic T cell response
Nature Immunology (2023)
-
Prevention, treatment and cure of HIV infection
Nature Reviews Microbiology (2023)
-
Neuroinflammatory transcriptional programs induced in rhesus pre-frontal cortex white matter during acute SHIV infection
Journal of Neuroinflammation (2022)
-
Alternative splicing and genetic variation of mhc-e: implications for rhesus cytomegalovirus-based vaccines
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.